Dimensionality Reduction
10/31/2022
Robert Utterback (based on slides by Andreas Muller)
Dimensionality Reduction
Principal Component Analysis
Principal Component Analysis
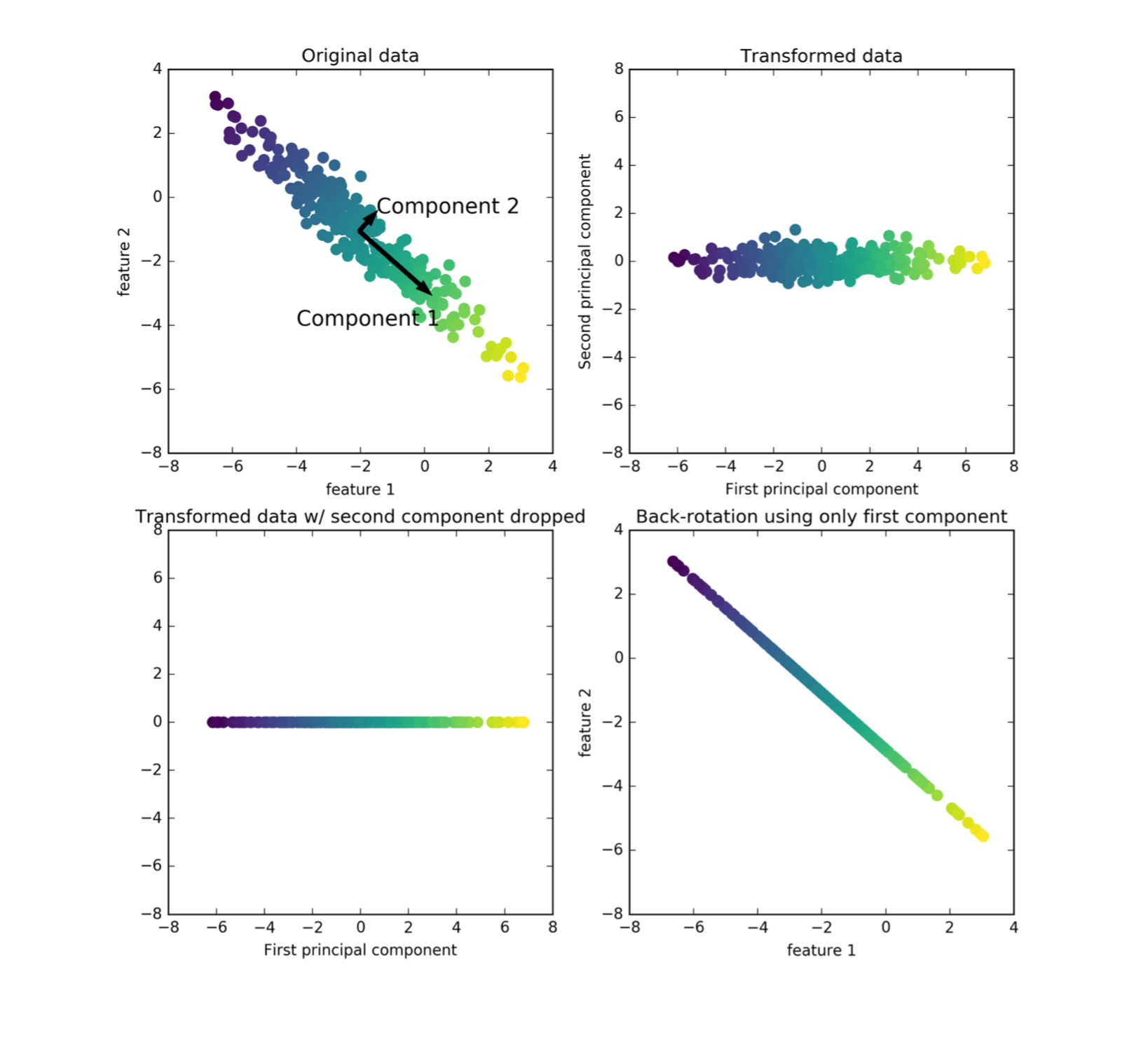
PCA objective(s)
\[\large\max\limits_{u_1 \in R^p, \| u_1 \| = 1} \text{var}(Xu_1)\] \[\large\max\limits_{u_1 \in R^p, \| u_1 \| = 1} u_1^T \text{cov} (X) u_1\]

PCA objective(s)
\[\large\min_{X', \text{rank}(X') = r}\|X-X'\|\]

PCA Computation
- Center X (subtract mean).
- In practice: Also scale to unit variance.
- Compute singular value decomposition:
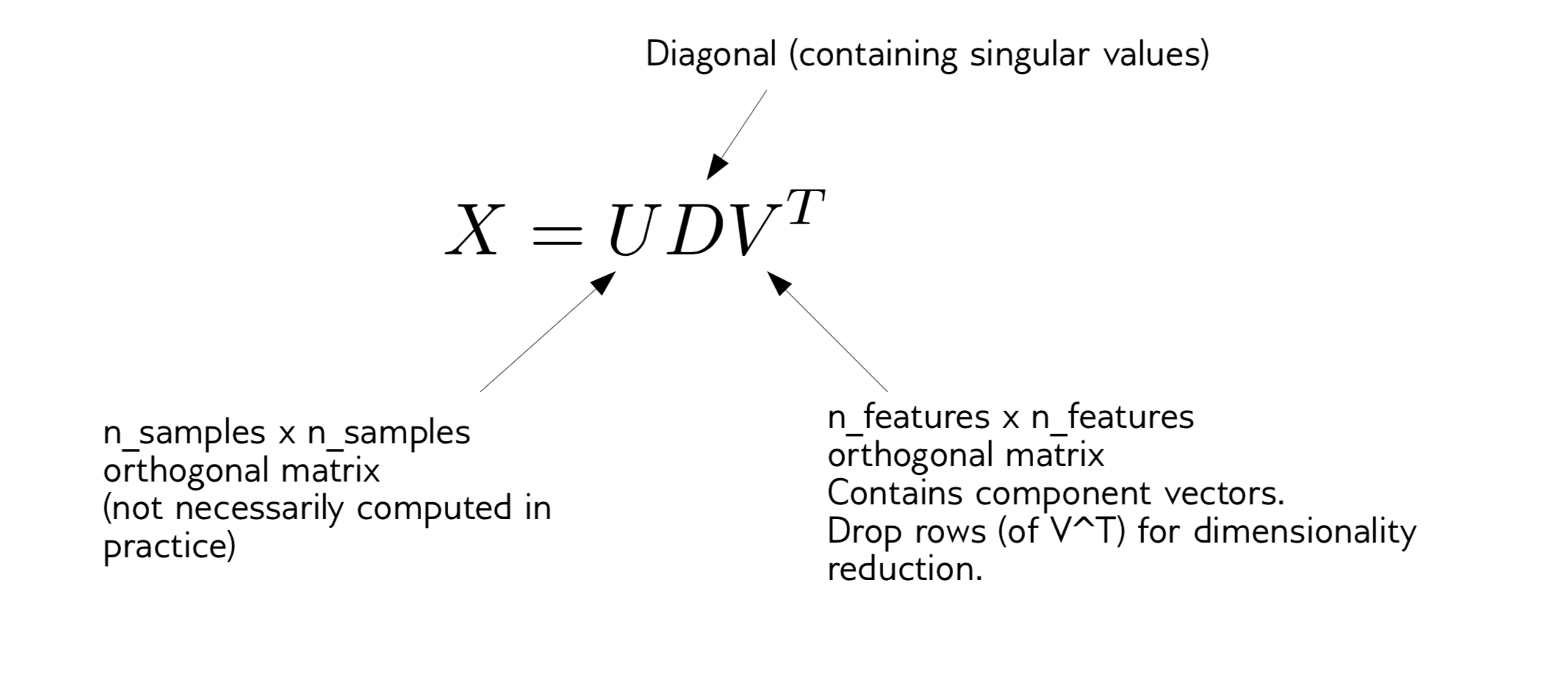
Whitening
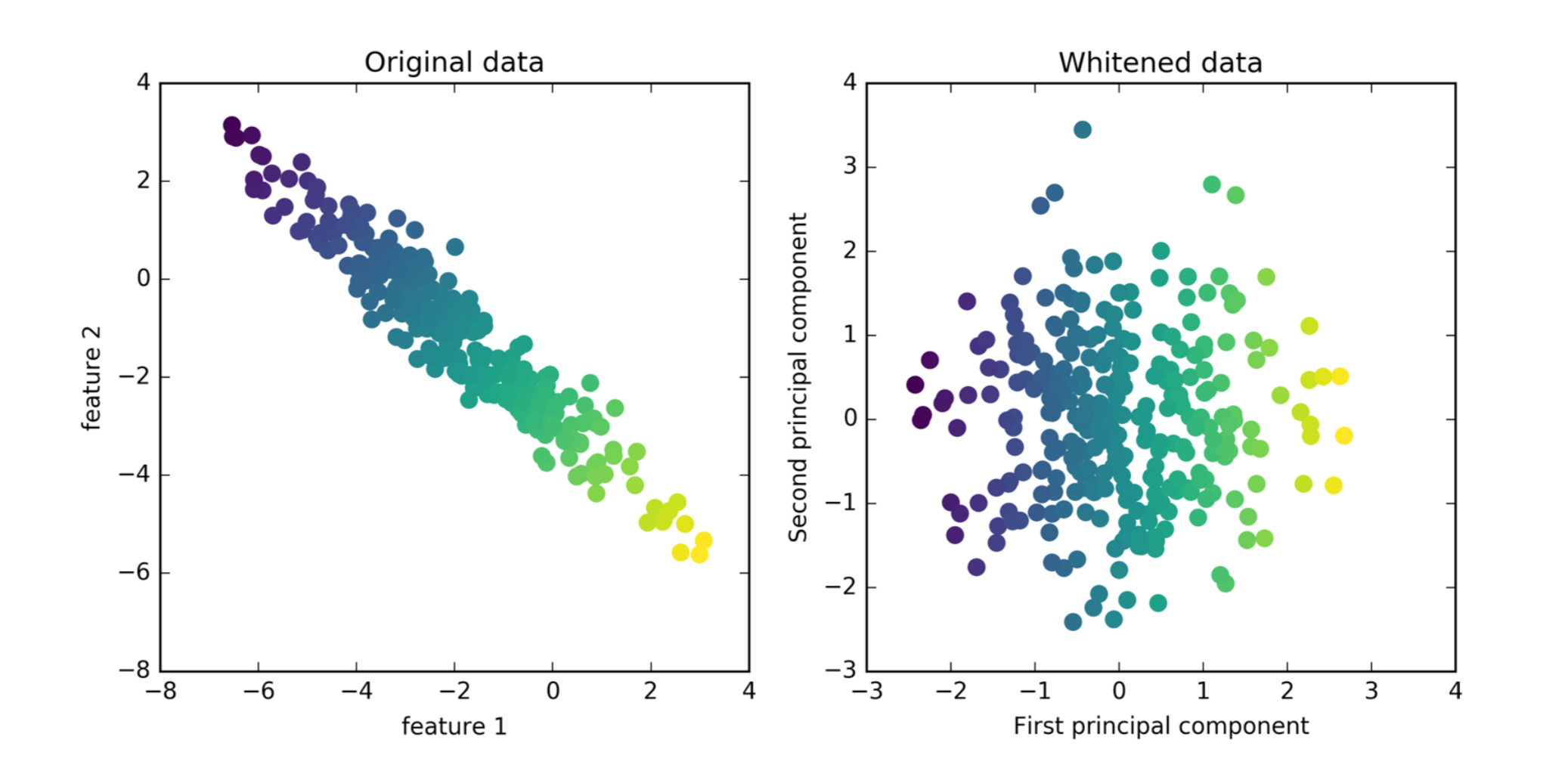
PCA for Visualization
PCA for Visualization
from sklearn.decomposition import PCA
print(cancer.data.shape)
pca = PCA(n_components=2)
X_pca = pca.fit_transform(cancer.data)
# print(X_pca.shape)
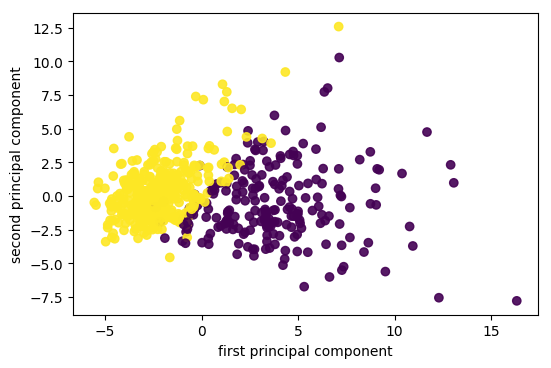
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=cancer.target)
plt.xlabel("first principal component")
plt.ylabel("second principal component")
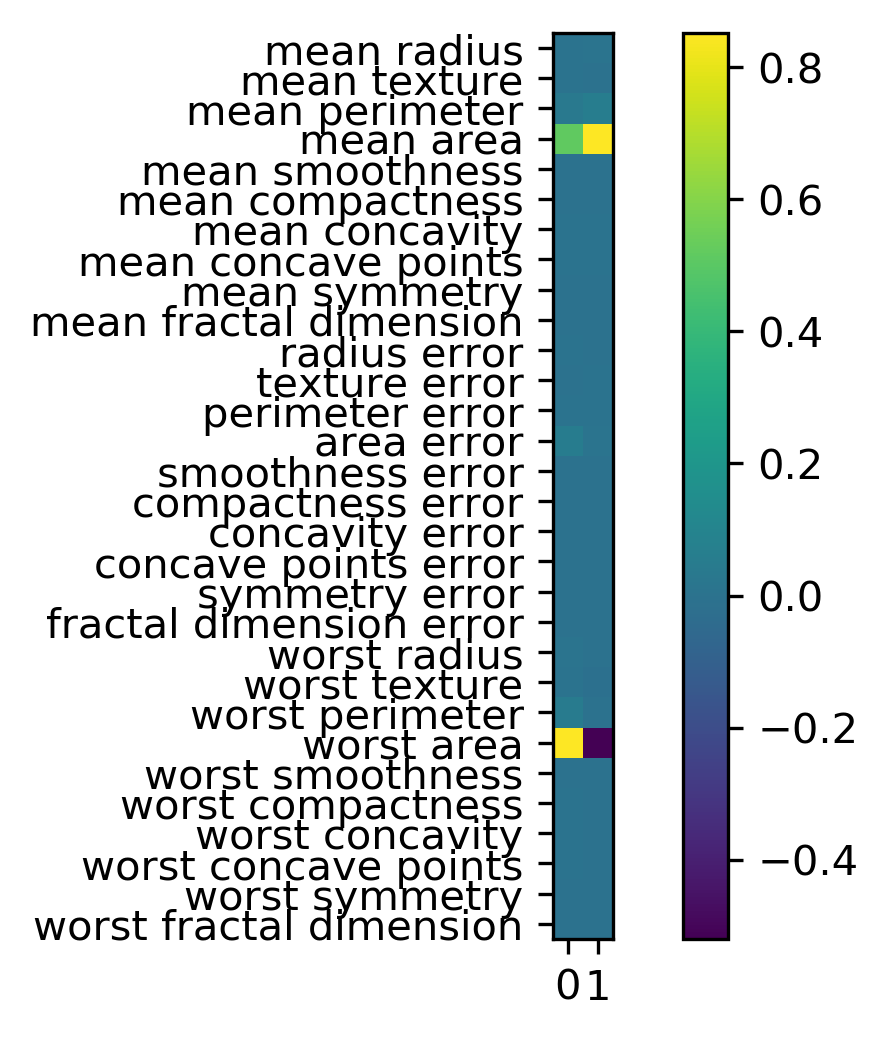
components = pca.components_
plt.imshow(components.T)
plt.yticks(range(len(cancer.feature_names)), cancer.feature_names)
plt.colorbar()
Scaling
pca_scaled = make_pipeline(StandardScaler(),
PCA(n_components=2),
LogisticRegression(C=10000))
pca_scaled.fit(X_train, y_train)
print(pca_scaled.score(X_train, y_train))
print(pca_scaled.score(X_test, y_test))
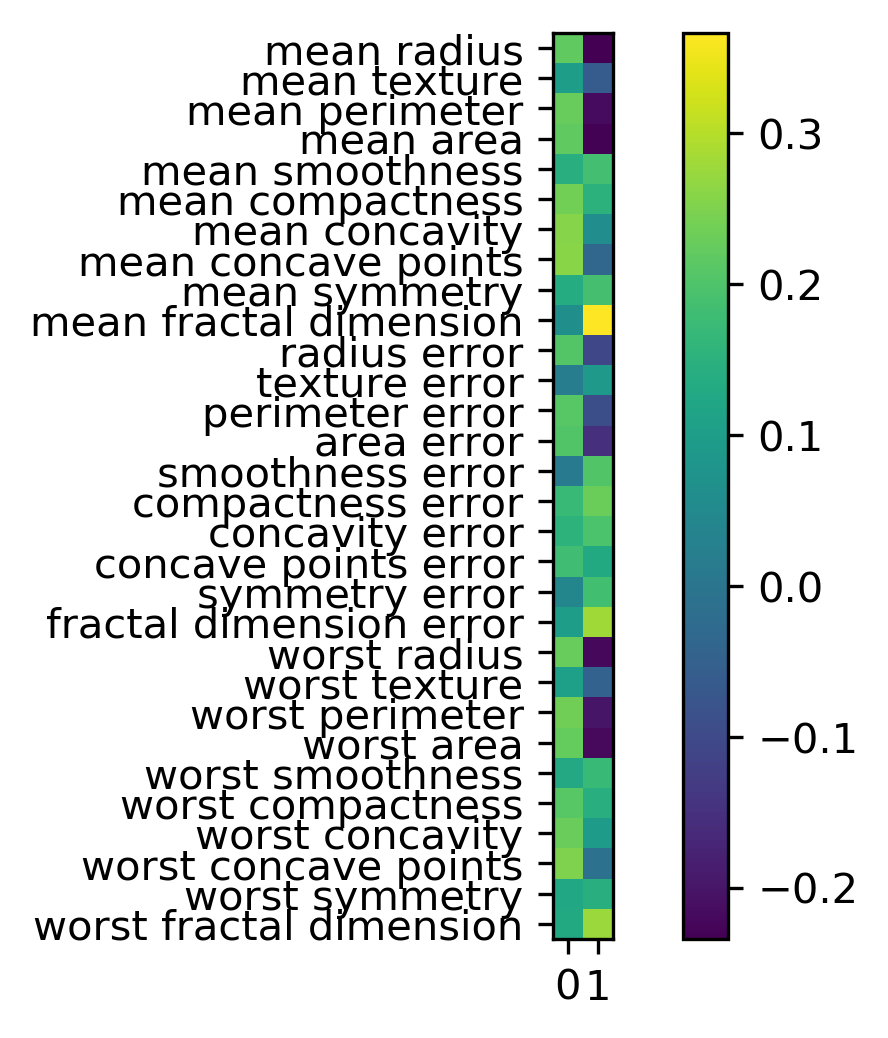
Inspecting Components
components = pca_scaled.named_steps['pca'].components_
plt.imshow(components.T)
plt.yticks(range(len(cancer.feature_names)),
cancer.feature_names)
plt.colorbar()

PCA for Regularization
PCA for Regularization
X_train, X_test, y_train, y_test = \
train_test_split(cancer.data, cancer.target,
stratify=cancer.target, random_state=0)
lr = LogisticRegression(C=10000, solver='liblinear')
lr.fit(X_train, y_train)
print(f"{lr.score(X_train, y_train):.3f}")
print(f"{lr.score(X_test, y_test):.3f}")
0.984 0.930
from sklearn.decomposition import PCA
lr = LogisticRegression(C=10000, solver='liblinear')
pca_lr = make_pipeline(StandardScaler(),
PCA(n_components=2), lr)
pca_lr.fit(X_train, y_train)
print(f"{pca_lr.score(X_train, y_train):.3f}")
print(f"{pca_lr.score(X_test, y_test):.3f}")
0.960 0.923
Variance Covered
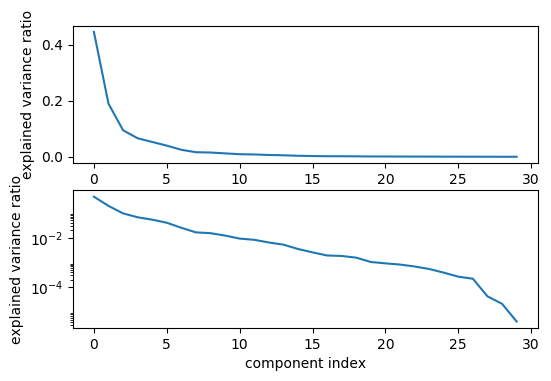
pca_lr = make_pipeline(StandardScaler(),
PCA(n_components=6), lr)
pca_lr.fit(X_train, y_train)
print(f"{pca_lr.score(X_train, y_train):.3f}")
print(f"{pca_lr.score(X_test, y_test):.3f}")
0.981 0.958
Interpreting Coefficients
pca = pca_lr.named_steps['pca']
lr = pca_lr.named_steps['logisticregression']
coef_pca = pca.inverse_transform(lr.coef_)
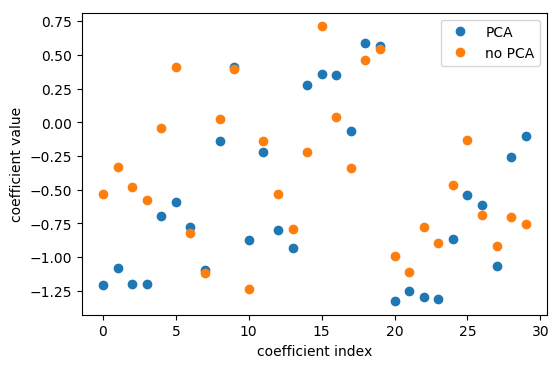
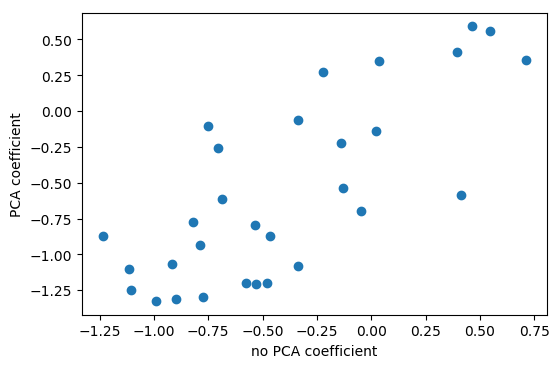
PCA is Unsupervised!
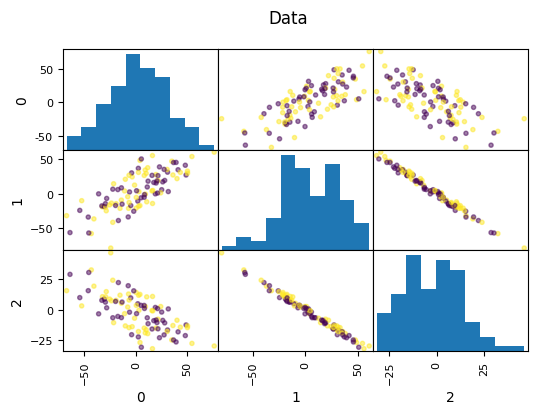
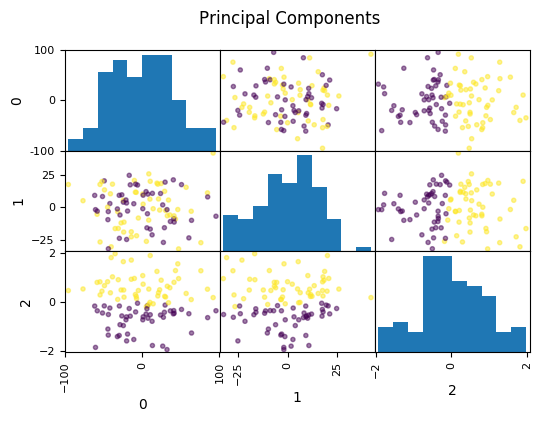
PCA for Feature Extraction
PCA for Feature Extraction
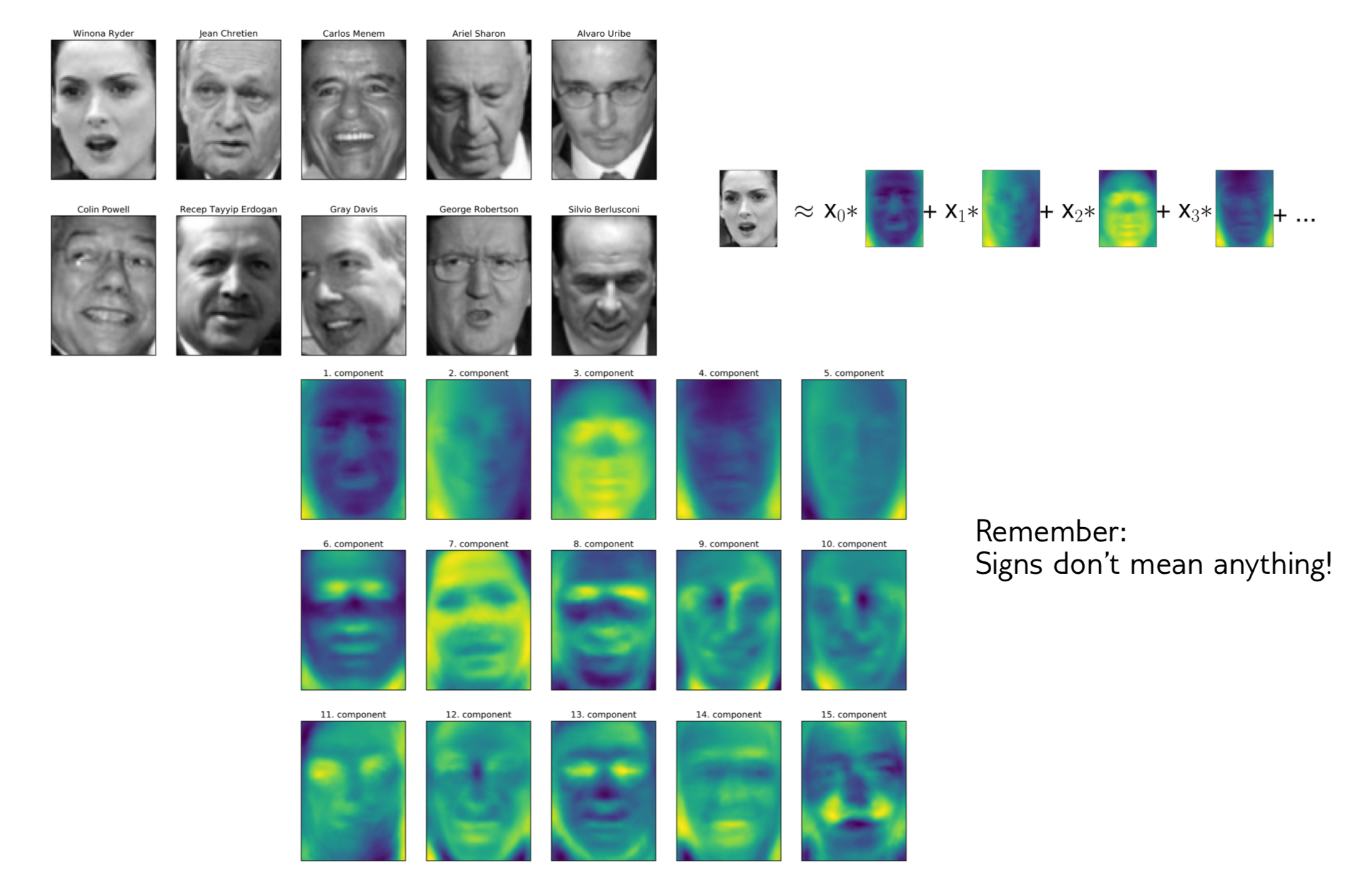
1-NN and Eigenfaces
from sklearn.datasets import fetch_lfw_people
from sklearn.neighbors import KNeighborsClassifier
people = fetch_lfw_people(min_faces_per_person=35, resize=0.7)
X_people, y_people = people.data, people.target
X_train, X_test, y_train, y_test = \
train_test_split(X_people, y_people,
stratify=y_people, random_state=0)
print(X_train.shape)
knn = KNeighborsClassifier(n_neighbors=1)
knn.fit(X_train, y_train)
print(f"{knn.score(X_test, y_test):.3f}")
(1539, 5655) 0.311
pca = PCA(n_components=100, whiten=True, random_state=0)
pca.fit(X_train)
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
print(X_train_pca.shape)
>>> (1539, 100)
knn = KNeighborsClassifier(n_neighbors=1)
knn.fit(X_train_pca, y_train)
print(f"{knn.score(X_test_pca, y_test):.3f}")
0.331
Reconstruction

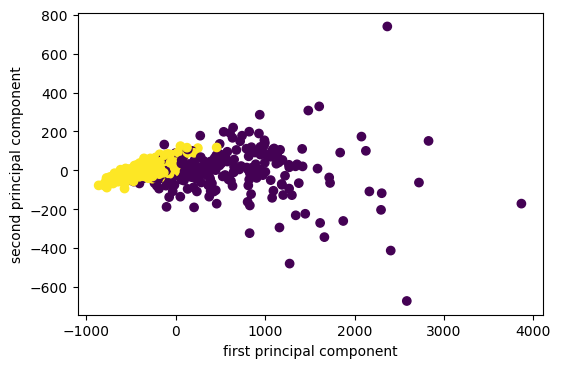
PCA for Outlier Detection
PCA for Outlier Detection
pca = PCA(n_components=100).fit(X_train)
inv = pca.inverse_transform(pca.transform(X_test))
reconstruction_errors = np.sum((X_test - inv)**2, axis=1)

Best reconstructions

Worst reconstructions
Manifold Learning
Manifold Learning
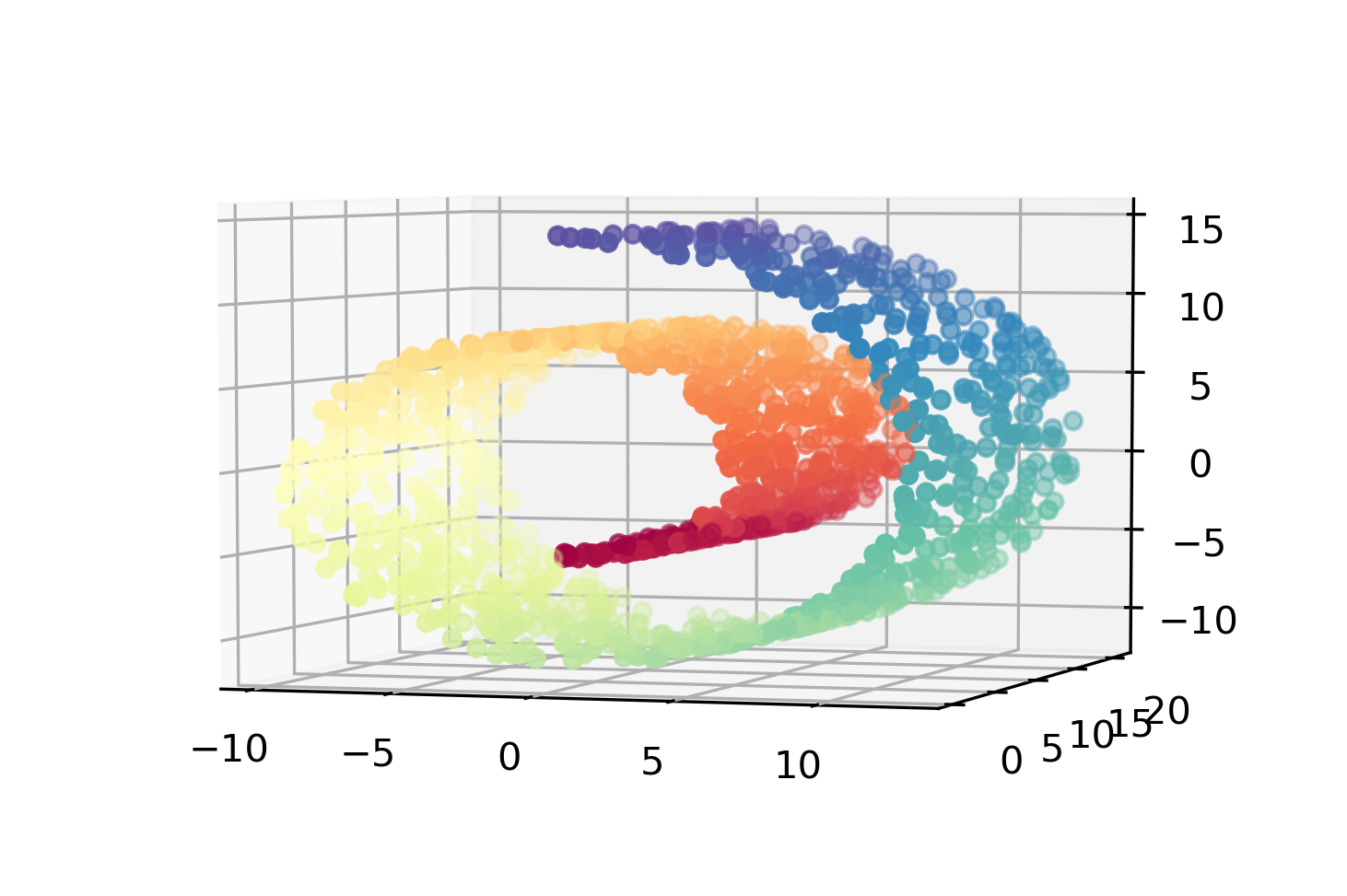
Pros and Cons
- For visualization only
- Axes don’t correspond to anything in the input space.
- Often can’t transform new data.
- Pretty pictures!
Algorithms in sklearn
KernelPCA– does PCA, but with kernels- Eigenvalues of kernel-matrix
- Spectral embedding (Laplacian Eigenmaps)
- Uses eigenvalues of graph Laplacian
- Locally Linear Embedding
- Isomap “kernel PCA on manifold”
- t-SNE (t-distributed stochastic neighbor embedding)
t-SNE
\[p_{j\mid i} = \frac{\exp(-\lVert\mathbf{x}_i - \mathbf{x}_j\rVert^2 / 2\sigma_i^2)}{\sum_{k \neq i} \exp(-\lVert\mathbf{x}_i - \mathbf{x}_k\rVert^2 / 2\sigma_i^2)}\]\[p_{ij} = \frac{p_{j\mid i} + p_{i\mid j}}{2N}\]
\[q_{ij} = \frac{(1 + \lVert \mathbf{y}_i - \mathbf{y}_j\rVert^2)^{-1}}{\sum_{k \neq i} (1 + \lVert \mathbf{y}_i - \mathbf{y}_k\rVert^2)^{-1}}\]
\[KL(P||Q) = \sum_{i \neq j} p_{ij} \log \frac{p_{ij}}{q_{ij}}\]
from sklearn.manifold import TSNE
from sklearn.datasets import load_digits
digits = load_digits()
X = digits.data / 16.
X_tsne = TSNE().fit_transform(X)
X_pca = PCA(n_components=2).fit_transform(X)
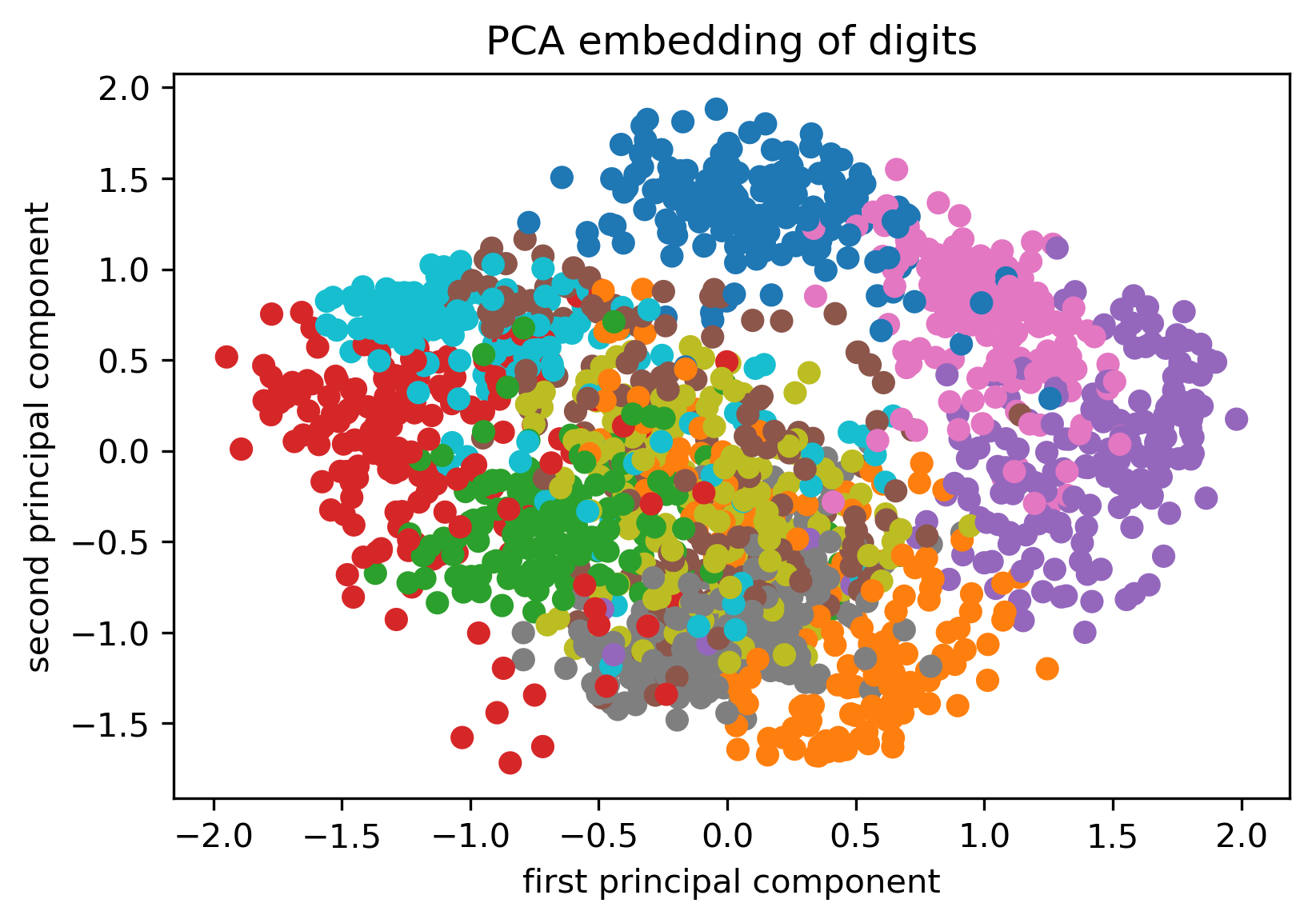
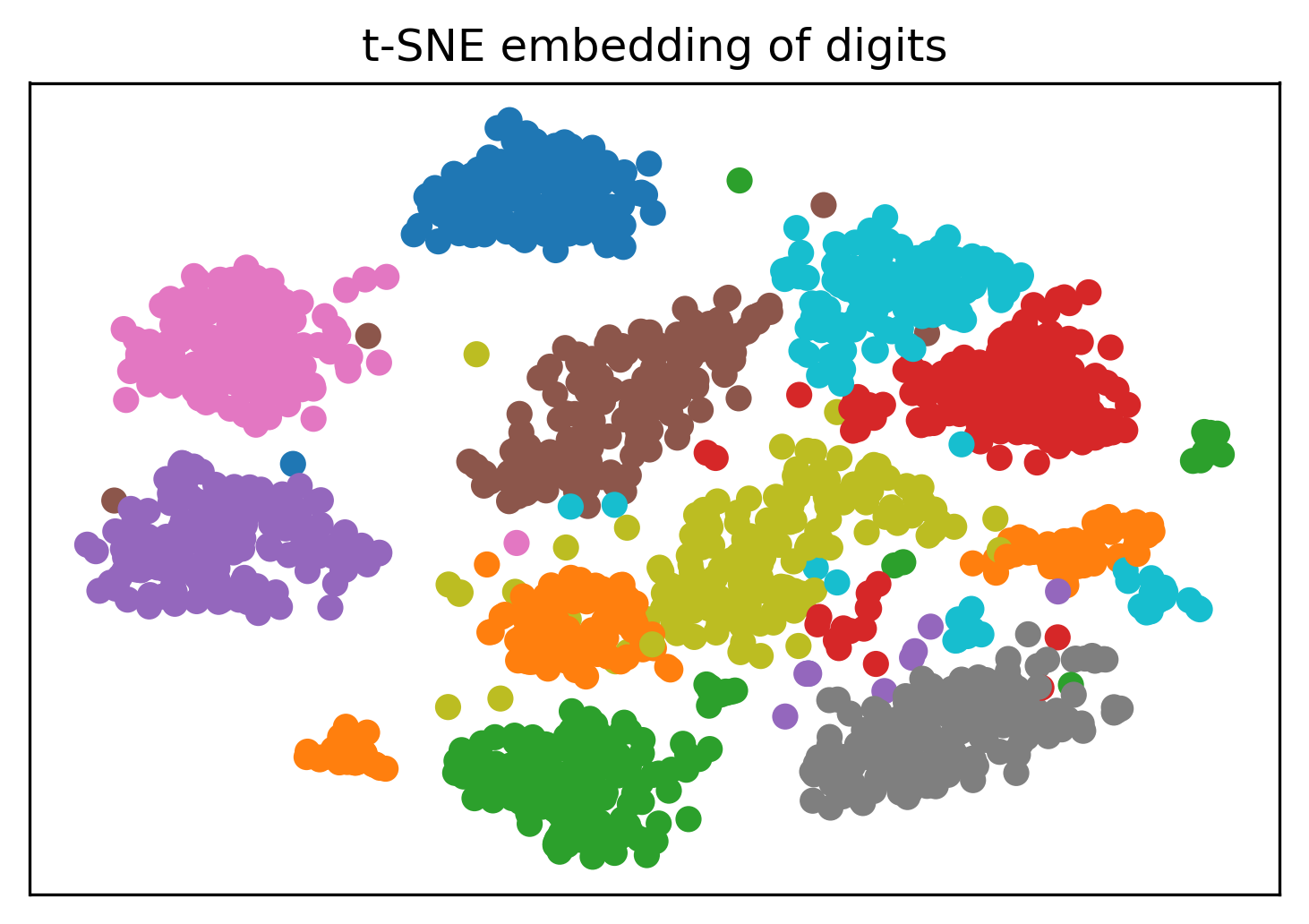
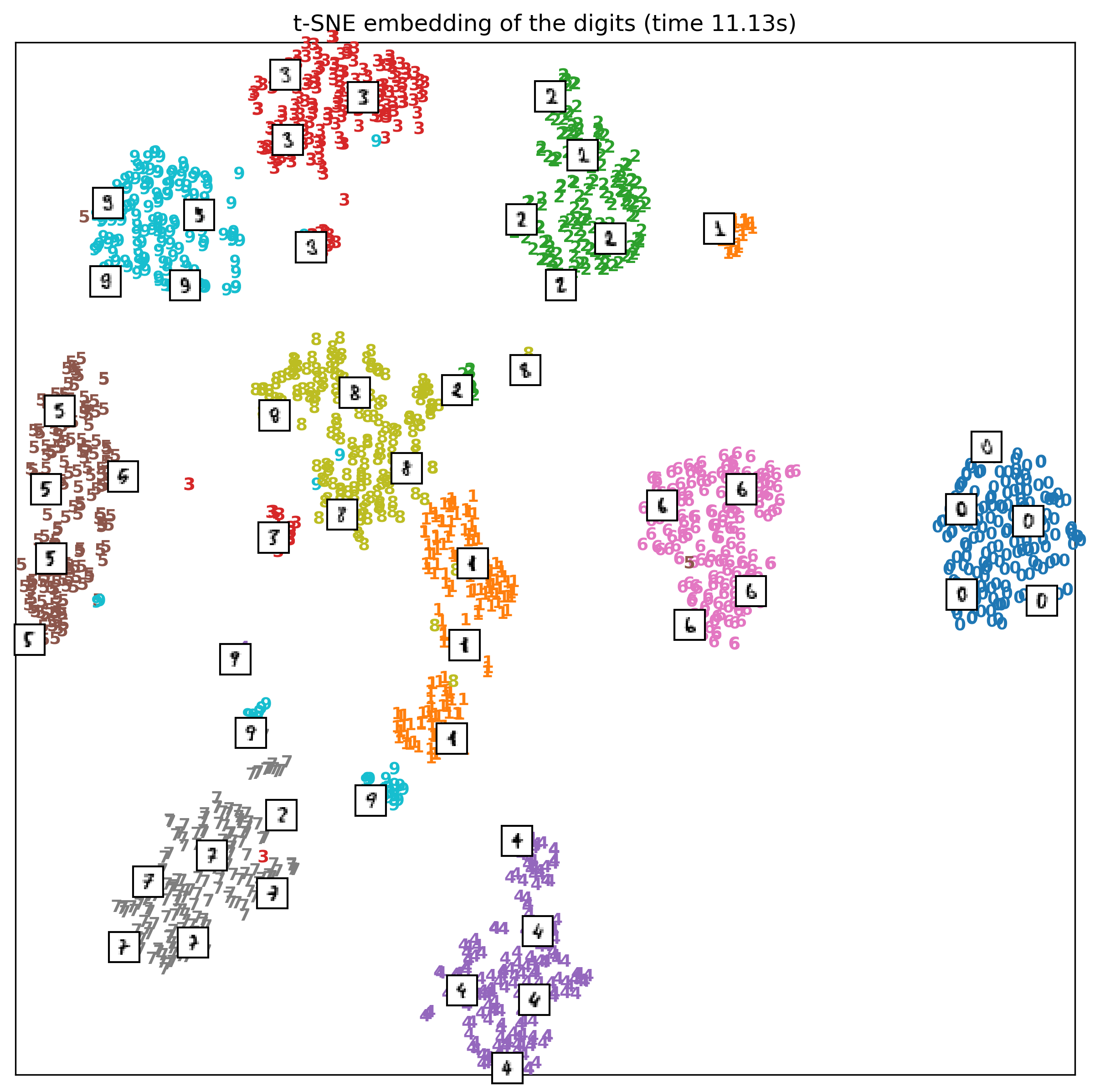
Tuning t-SNE perplexity
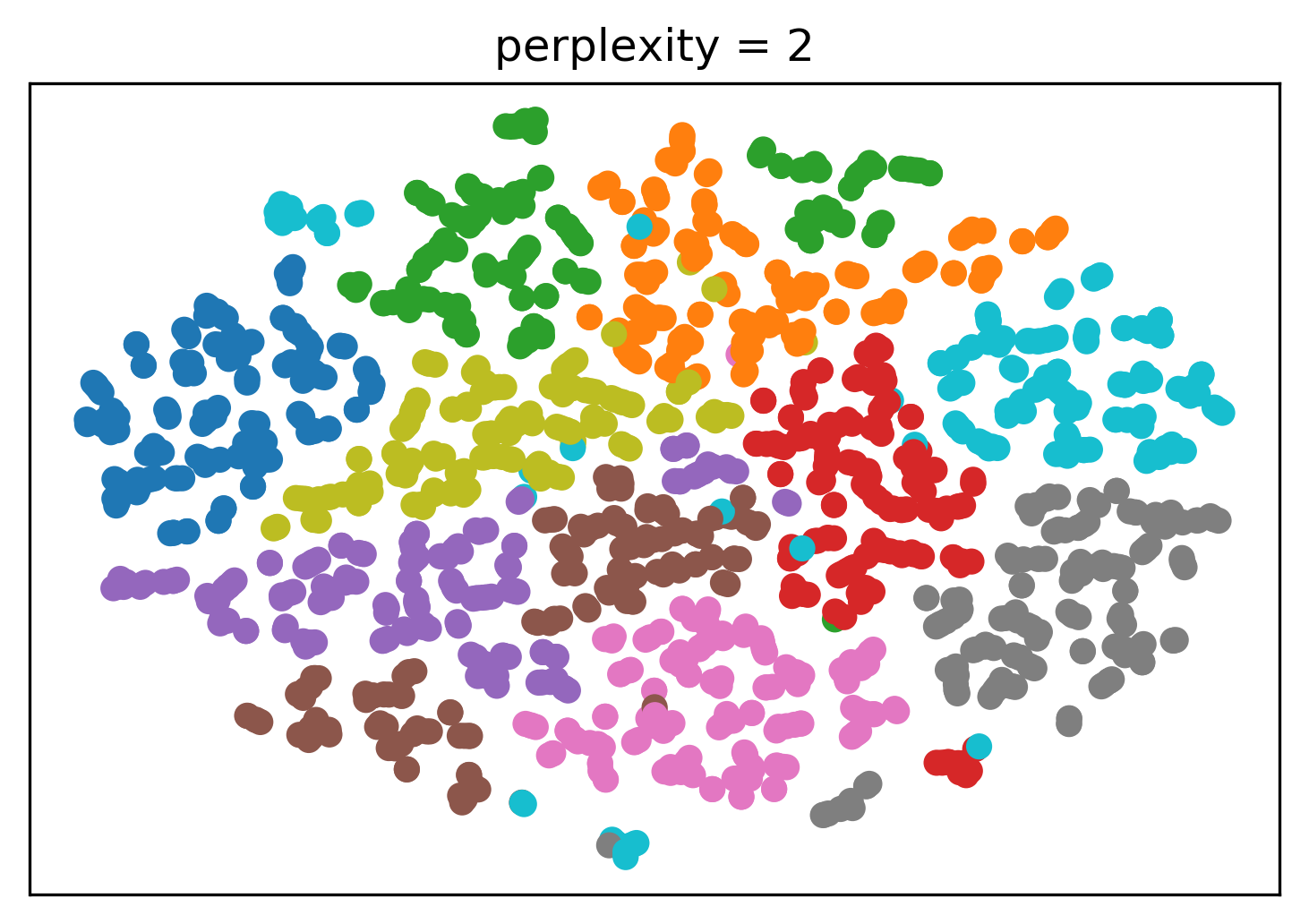
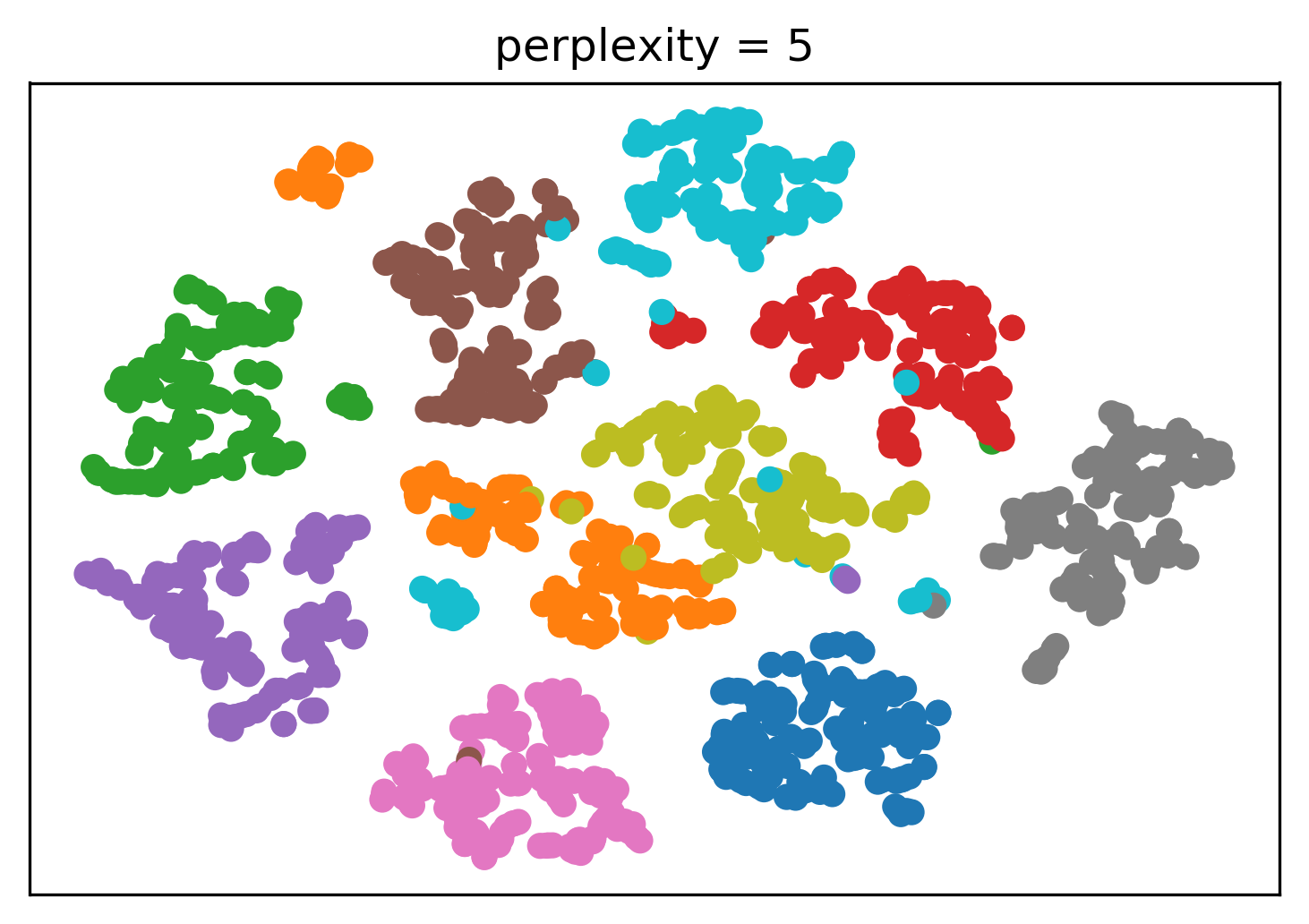
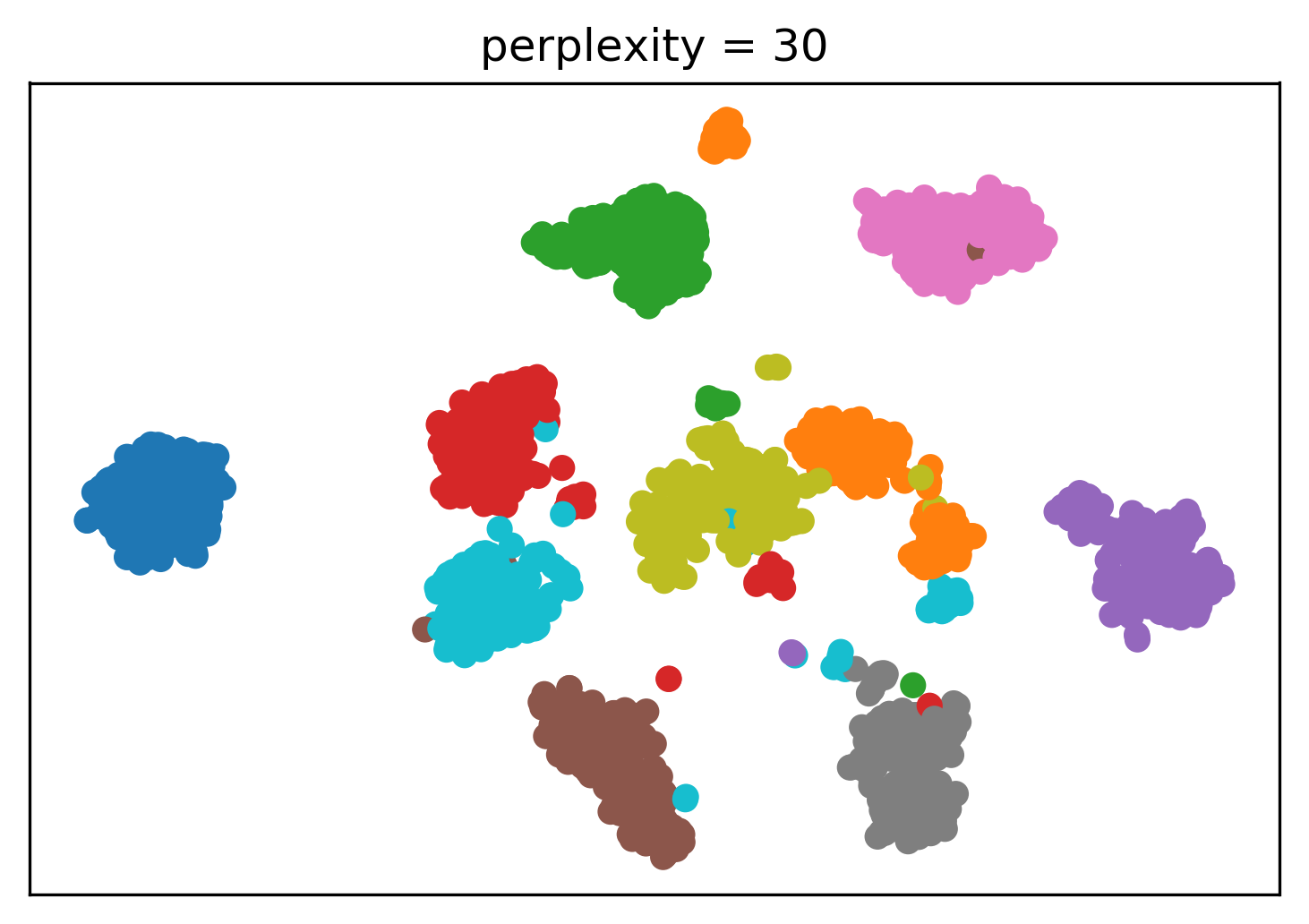
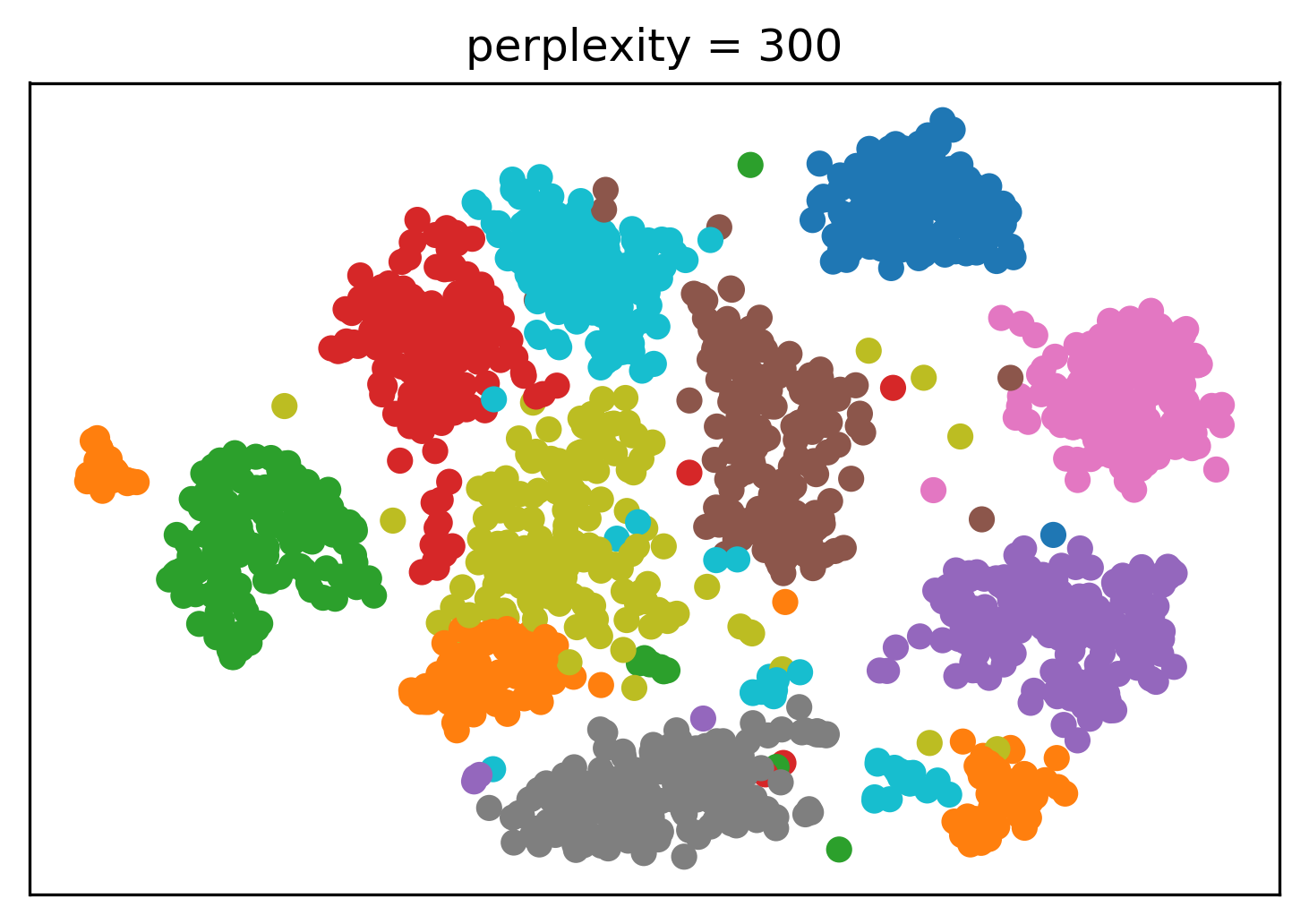
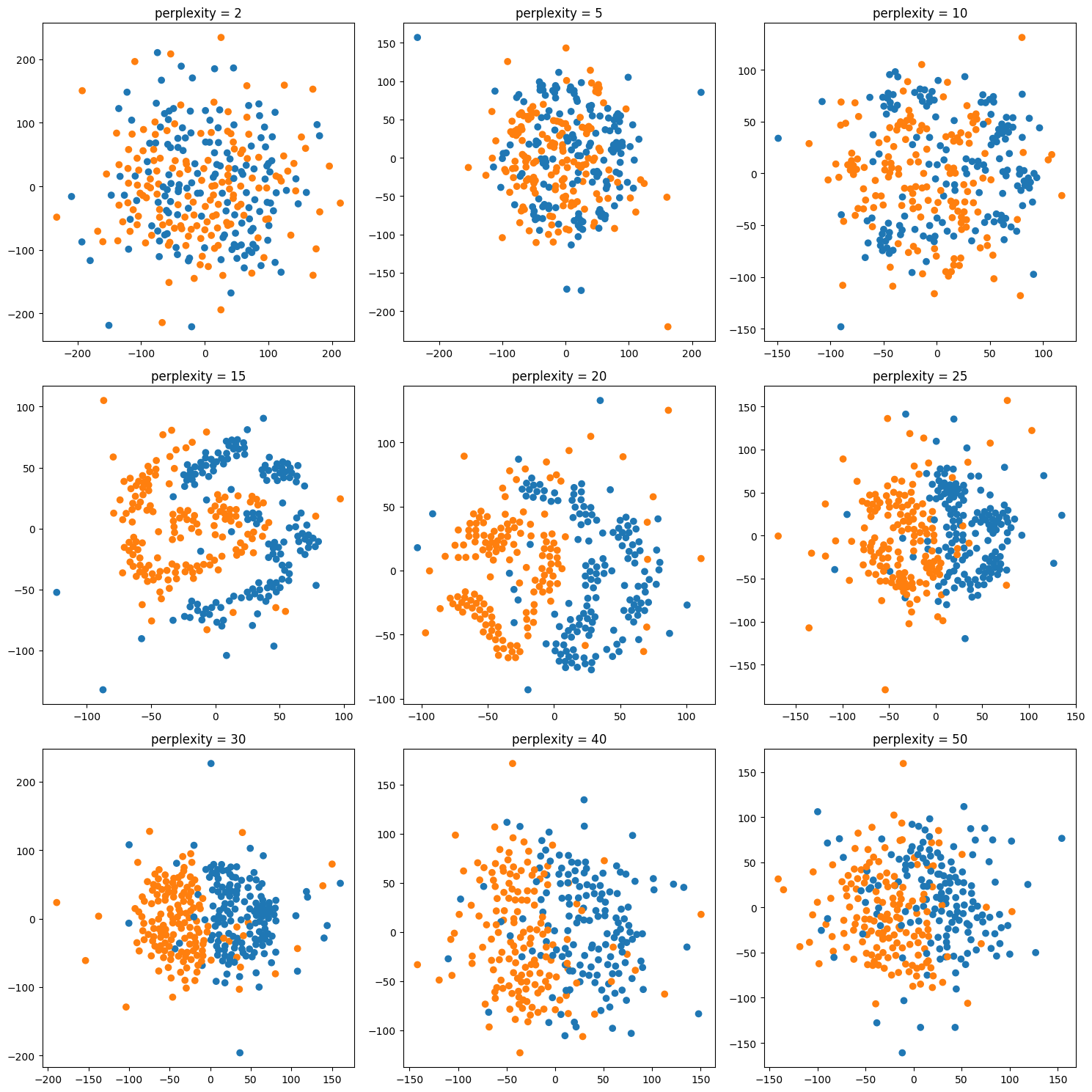
Play around online
Discriminant Analysis
Linear Discriminant Analysis aka Fisher Discriminant
\[ P(y=k | X) = \frac{P(X | y=k) P(y=k)}{P(X)} = \frac{P(X | y=k) P(y = k)}{ \sum_{l} P(X | y=l) \cdot P(y=l)}\]
\[ p(X | y=k) = \frac{1}{(2\pi)^n |\Sigma|^{1/2}}\exp\left(-\frac{1}{2} (X-\mu_k)^t \Sigma^{-1} (X-\mu_k)\right) \]
\[ \log\left(\frac{P(y=k|X)}{P(y=l | X)}\right) = 0 \Leftrightarrow (\mu_k-\mu_l)\Sigma^{-1} X = \frac{1}{2} (\mu_k^t \Sigma^{-1} \mu_k - \mu_l^t \Sigma^{-1} \mu_l) \]
Quadratic Discriminant Analysis
\[ p(X | y=k) = \frac{1}{(2\pi)^n |\Sigma_k|^{1/2}}\exp\left(-\frac{1}{2} (X-\mu_k)^t \Sigma_k^{-1} (X-\mu_k)\right) \]
Comparison
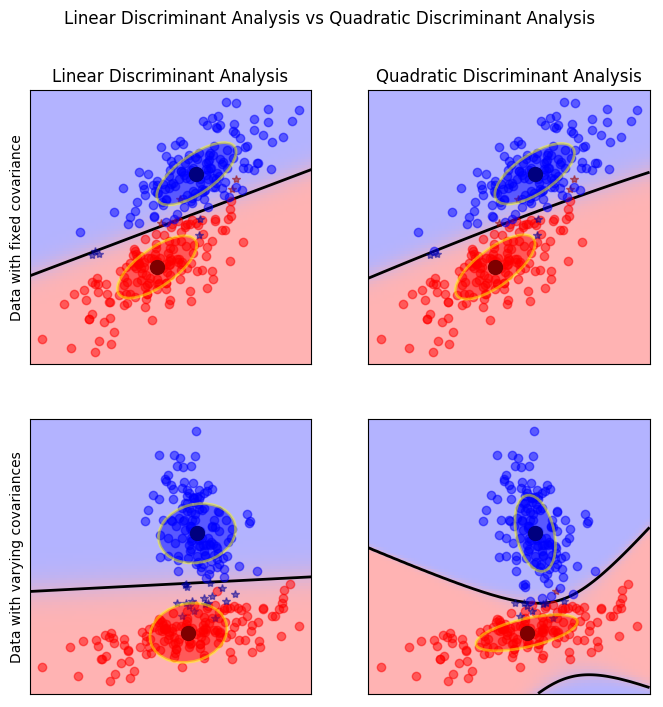
Discriminants and PCA
- Both fit Gaussian model
- PCA for the whole data
- LDA multiple Gaussians with shared covariance
- Can use LDA to transform space
- At most as many components as there are classes - 1 (needs between-class variance)
PCA vs Linear Discriminants
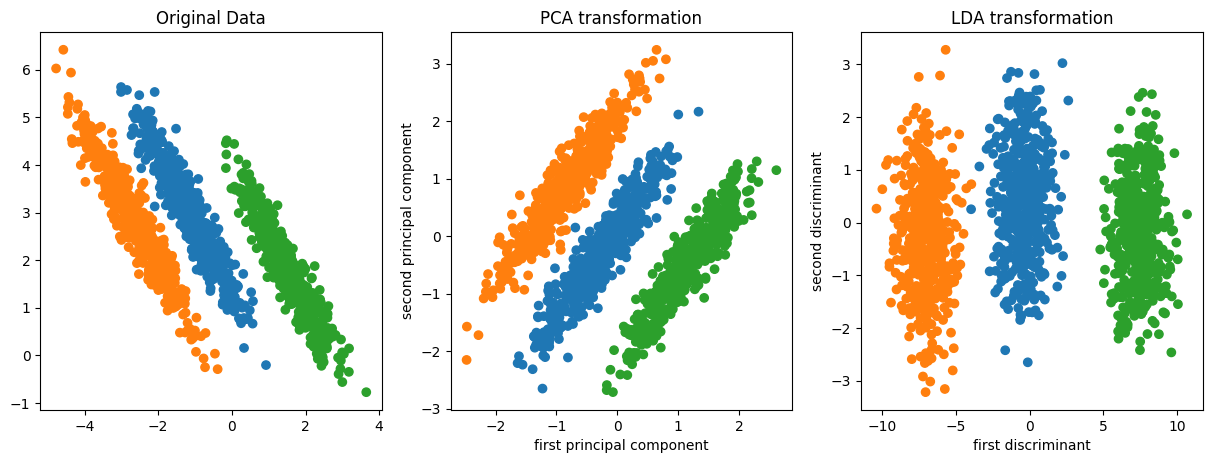
Data where PCA failed
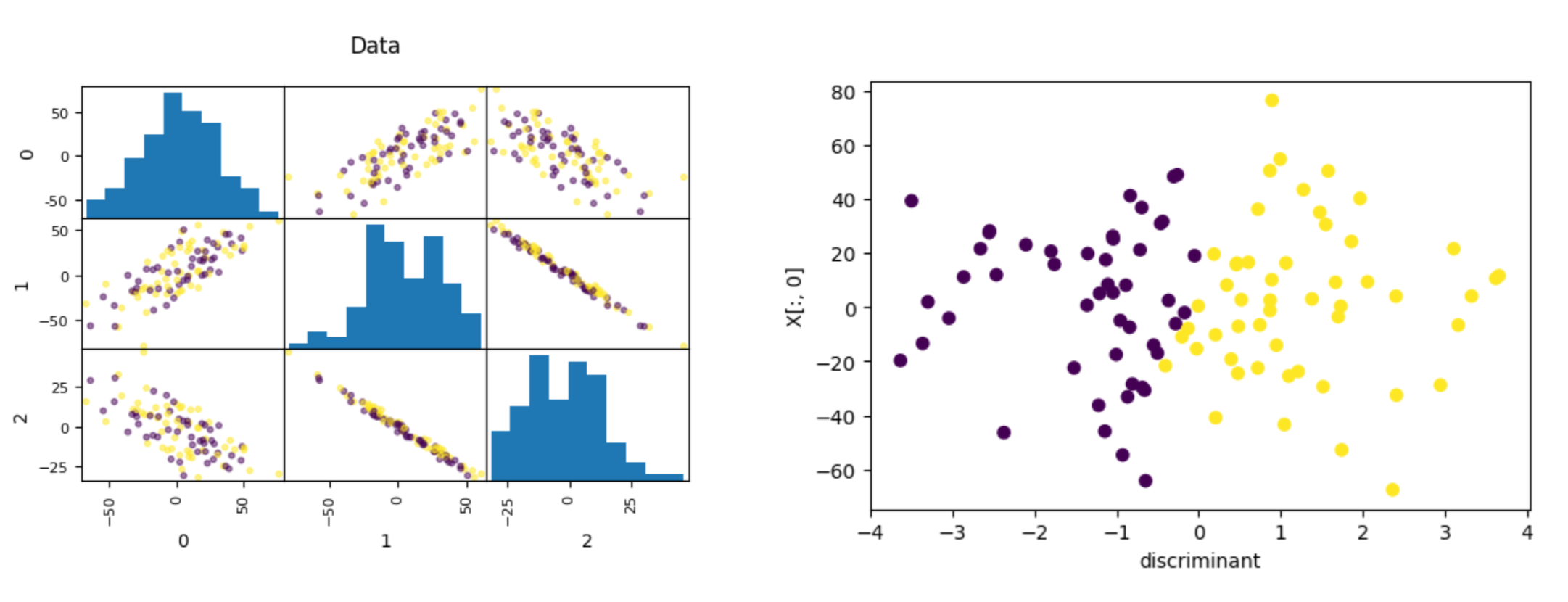
Summary
- PCA good for visualization, exploring correlations
- PCA can sometimes help with classification as regularization or for feature extraction
- Manifold learning makes nice pictures
- LDA is a supervised alternative to PCA